Optimizing and characterizing bat intestinal organoids
Intestinal organoids were derived from horseshoe bats with a perfect establishment rate as we reported previously24 and consecutively passaged for around 3 months when cultured in the medium supplemented with a Wnt3a conditioned medium. We optimized the composition of the culture medium to lengthen the expandability of bat intestinal organoids and have a greater amount of stable bat organoids for experimentation and reduce the usage of live bats. A next-generation surrogate Wnt agonist was reported to support the growth of human intestinal organoids more efficiently than the Wnt3a conditioned medium.28 Thus, we switched to a modified culture medium (Supplementary Table 1) in which the Wnt surrogate replaced the original Wnt3a conditioned medium. The addition of the Wnt surrogate sustained a dramatically prolonged expansion of bat intestinal organoids for over one year. We routinely maintained 3 lines of bat organoids derived from 3 horseshoe bats (Rhinolophus sinicus) and passaged these organoids every 7 days for daily experimentation (Fig. 1a). Transmission electron microscopy revealed that the bat intestinal organoids consisted of enterocyte, goblet cell, Paneth cell and enteroendocrine cell (Fig. 1b); goblet cell, especially enterocyte, were the dominant cell populations in bat intestinal organoids (the bottom right panel in Fig. 1b), consistent with the many prior studies in human intestinal organoids.19,24,26 Due to the lack of specific antibodies against bat proteins, we used cell-type specific antibodies against human analogs. Immunostaining using human cell-type specific antibodies verified the presence of MUC2 + goblet cells and abundant Villin+ enterocytes in bat intestinal organoids (Fig. 1c). Thus, the optimized bat intestinal organoids are long-term expandable and faithfully simulate the bat intestinal epithelium. The comparable cellular composition of bat and human intestinal organoids provided a unique and biologically-active model system, enabling an analogous comparison of bat and human intestinal epithelial cells in vitro.
Fig. 1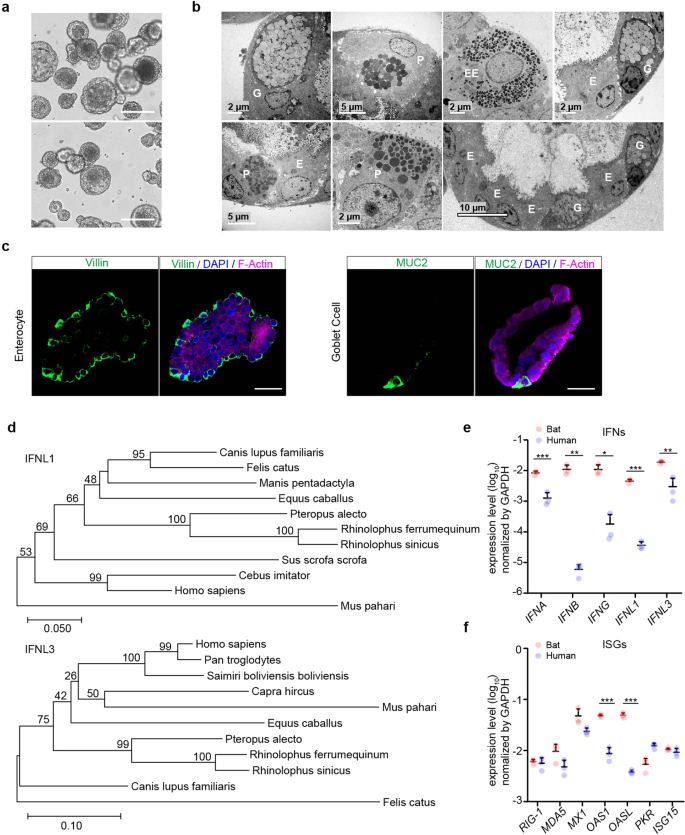
Characterization of optimized bat intestinal organoids and detection of the basal expression of antiviral genes. a Photomicrographs of optimized bat intestinal organoids on day 7 after passaging (magnification ×100). Scale bar, 100 μm. b Transmission electron microscopy illustrates the ultrastructural morphology of absorptive enterocyte (E), Paneath cell (P), goblet cell (G), and enteroendocrine cell (EE) in bat intestinal organoids. c Bat intestinal organoids were fixed and immunostained to label Villin + (green) enterocyte and MUC2 + (green) goblet cell. Nuclei and actin filaments were counterstained with DAPI (blue) and Phalloidin-647 (purple), respectively. Scale bar, 20 µm. d Phylogenetic analysis based on an alignment of horseshoe bat (Rhinolophus sinicus) IFNL1 and IFNL3 ORF cDNAs with other Chiroptera species and mammals. Bootstrap values (%) are indicated on each branch, and the scale for branch length is shown at the bottom of the tree. e, f GAPDH-normalized expression levels of IFN genes (e) and ISGs (f) in bat and human intestinal organoids. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3. Two-tailed unpaired Student’s t test
Detecting the basal expression of antiviral genes in bat intestinal organoids
Type III interferons (IFN) are the major player of mucosal immunity in the intestines of human and mouse.29 IFN lambda-1 (IFNL1) and IFNL3 were highly induced in human intestinal organoids upon viral infections.21,26 We first determined the sequences of horseshoe bat type III IFNs using RACE PCR. A long coverage genome sequence of the Chinese horseshoe bat (Rhinolophus sinicus) is publicly available in the GenBank Bioproject database (Accession No. PRJNA294852). Sequences of analogous human genes, including IFNL1 and IFNL3, were blast searched in the genome of Chinese horseshoe bat. The matched sequences were used to design RACE PCR primers (Supplementary Table 2) to detect partial sequences. Then 5’ and 3’ RACE PCR assay was performed using RNA extracted from bat intestinal organoids. We obtained two sequences with high homology to interferon lambda-1 like and interferon lambda-3 like genes in the closely related bat species Rhinolophus ferrumenguinum, which were designated IFNL1 and IFNL3, respectively. A comparison of the identified IFNLs with other mammals demonstrated that IFNL1 and IFNL3 of Rhinolophus sinicus were identical to those of Rhinolophus ferrumequinum and 99% homologous to those of Pteropus alecto (Fig. 1d).
We also designed qPCR primers using the same approach and examined genes of innate immunity in bat intestinal organoids. We examined the basal levels of IFNs and several reported ISGs15 in bat intestinal organoids and compared them with those in the human organoids that were cultured with the same protocol (Fig. 1e). We found that bat organoids expressed all three types of IFNs with 1–3 log units higher than human organoids. Several ISGs such as OAS1 and OASL were approximately 10-fold higher in bat organoids than their human counterparts, while others, such as MX1, and MDA5 that were IFN-inducible in bat cells,16 tended to be expressed higher in bat organoids (Fig. 1f). Overall, basal expression levels of IFNs and some ISGs were significantly higher in bat intestinal organoids than in their human counterparts.
Bat organoids eliciting a more robust antiviral response to Poly(I:C) treatment compared to human organoids
We then analyzed the induction of antiviral response in bat and human intestinal organoids. Poly(I:C), a synthetic virus mimic, induced innate immunity in human intestinal cells and intestinal organoids through binding to TLR3 and MDA5,30 pattern recognition receptors on human intestinal cells.31 First, we tested the conditions to deliver Poly(I:C) to bat intestinal organoids for a robust induction of cellular response. We found that incubation of bat organoids with 10 µg/ml Poly(I:C) after mechanical shearing consistently and effectively induced antiviral genes (Supplementary Fig. 1). We then incubated bat and human intestinal organoids in parallel with or without 10 µg/ml Poly(I:C) after shearing and examined expression levels of antiviral genes in bat and human intestinal organoids relative to those in mock-treated organoids. Type III IFNs, especially IFNL1, were highly induced in bat organoids after Poly(I:C) stimulation, whereas type I and II IFNs, including IFN-α (IFNA), -β (IFNB) and -γ (IFNG), were modestly induced, with a much lower magnitude than that of IFNL1 and IFNL3 (Fig. 2a), very consistent with the previous findings of type III IFNs as a major player in mucosal immunity of human and mouse.21,26 Several ISGs, including ISG15, MX1 and MDA5, were highly upregulated with a slight delay compared to IFNs (Fig. 2a). Human intestinal organoids displayed a similar profile of immune activation. However, the induction of type III IFNs and ISGs, was more rapid and potent, and sustained longer in bat organoids than in their human counterparts (Fig. 2b). More active and prolonged induction of antiviral defense in bat organoids was observed in multiple experiments comparing bat and human intestinal organoids from different donors (Supplementary Fig. 2). The secretion of human IFNL1 and IFNL3 from Poly(I:C)-treated human intestinal organoids was verified by ELISA (Fig. 2c), and Poly(I:C)-induced upregulation of human ISG15 was shown by Western blotting (Fig. 2d and Supplementary Fig. 6). The higher magnitude of innate immune activation in bat organoids than in human organoids was intrinsic since bat and human organoids showed a comparable uptake of fluorescein-labeled Poly(I:C) as determined by flow cytometry analysis. The proportion of fluorescein-positive cells and the fluorescence intensity were comparable in bat and human intestinal organoids after incubation with fluorescein-labeled Poly(I:C) (Fig. 2e, f and Supplementary Figs. 3 and 6).
Fig. 2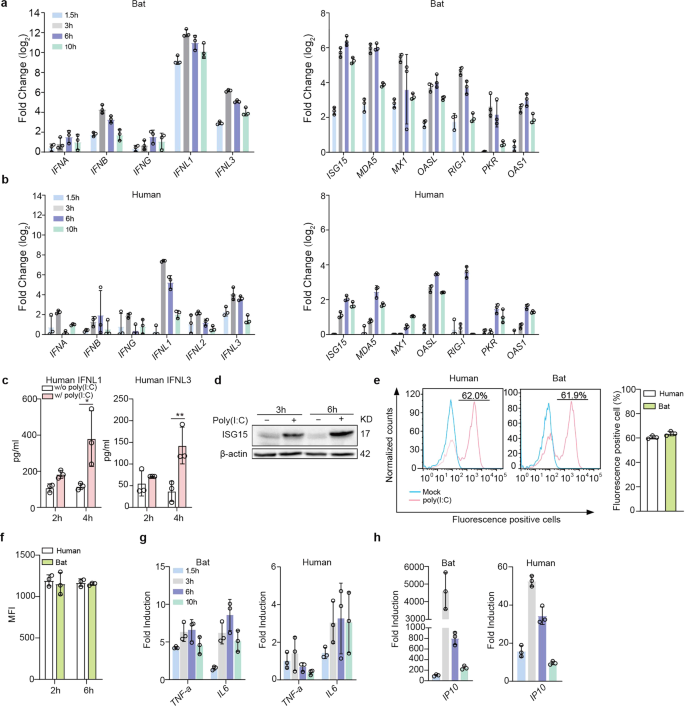
Bat organoids elicited a more robust antiviral response to Poly(I:C) treatment. Bat and human intestinal organoids were sheared mechanically and treated with 10 μg/ml Poly(I:C) or mock-treated with DMSO. a, b Induction of IFNs (left) and ISGs (right) in bat intestinal organoids (a) and human intestinal organoids (b) at the indicated hours after Poly(I:C) treatment. Results show the log2-fold change of GAPDH-normalized expression level in the treated organoids relative to mock-treated organoids. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3. c Culture media from the treated or mock-treated human intestinal organoids at the indicated hours were applied to ELISA to measure concentrations of IFNL1 and IFNL3. Data represent the mean and s.d. of a representative experiment in one line of human organoids, n = 3. Two-tailed unpaired Student’s t test. d Poly(I:C) treated or mock-treated human intestinal organoids were collected at the indicated hours post-treatment and subjected to Western blot to detect human ISG15. e, f Bat and human intestinal organoids were sheared and incubated with 10 μg/ml Poly(I:C) Fluorescein in triplicate for 2 h and 6 h. The organoids were then dissociated and applied to flow cytometry to detect the percentage at 2 h post treatment (e) and mean fluorescence intensity (MFI, f) of Fluorescein-positive cells at 2 and 6 h post treatment. g, h Induction of TNF-a and IL6 (g) and IP10 (h) in bat and human intestinal organoids at the indicated hours after treatment. Results show the fold change of GAPDH-normalized expression level in Poly(I:C)-treated organoids relative to mock-treated organoids. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3
Poly(I:C) is a dual agonist of TLR3 and MDA5.30 Apart from IFN induction, TLR3 or MDA5 activation also leads to the production of proinflammatory cytokines through NF-κB signaling. We examined proinflammatory cytokines and found that IL6 and TNF-α were induced in bat intestinal organoids (left panel, Fig. 2g). Notably, IP10 was rapidly and dramatically induced after Poly(I:C) treatment (left panel, Fig. 2h). Human intestinal organoids showed similar induction kinetics, yet with a much lower magnitude than bat organoids (right panels, Fig. 2g, h).
TLR3 and RLR signaling pathways mediating antiviral response in bat and human organoids
We proceeded to elucidate signaling pathway(s) mediating immune activation in bat organoids. As aforementioned, Poly(I:C) is a synthetic agonist of TLR3 and MDA5. BX795 is a catalytic inhibitor of TBK1/IKKε, a kinase in TLR3 and RLR pathways. CYT387, a potent inhibitor of TBK1/IKKε, can also inhibit JAK1 and JAK2,32 Janus kinases in the JAK/STAT pathway mediating IFN signaling. We first determined the appropriate concentrations of the two inhibitors to exclude the potential artifact of cytotoxicity confounding the readouts in bat and human organoids (Supplementary Fig. 4). Bat and human intestinal organoids were pretreated with BX795 or CYT387 or DMSO overnight with concentrations of minimal cytotoxicity, followed by Poly(I:C) stimulation or mock stimulation and further incubation with the initial concentrations of BX795 or CYT387 or DMSO (Fig. 3a). We then examined IFNL1 and IFNL3 induction in the organoids at 2 and 4 h after Poly(I:C) stimulation relative to those with mock stimulation. BX795 inhibition suppressed Poly(I:C) induced bat IFNL1 and IFNL3 in a dose-dependent manner (Fig. 3b). CYT387 appeared to have a more potent inhibitory effect on Poly(I:C)-triggered induction of IFNL1 and IFNL3 in bat organoids (Fig. 3c). Similarly, BX795 (Fig. 3d) and CYT387 (Fig. 3e) treatment abrogated Poly(I:C)-stimulated IFNL1 and IFNL3 upregulation in human intestinal organoids. Again, Poly(I:C)-stimulated IFNL1 and IFNL3 production less intensively in human organoids than in bat intestinal organoids. In addition, Poly(I:C)-triggered IFNL1 (Fig. 3f) and IFNL3 (Fig. 3g) secretion from human organoids decreased in a dose-dependent manner with increasing concentrations of BX795 and CYT387.
Fig. 3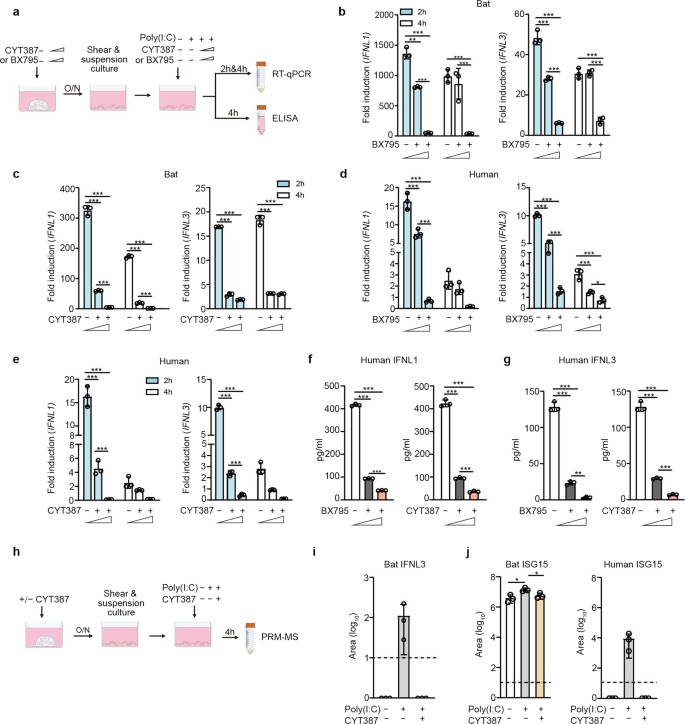
TLR3 and RLR signaling pathways mediate antiviral responses in bat and human organoids. a A schematic graph outlines the experimental procedure for panels b–g. Bat and human intestinal organoids were pretreated with BX795 (0, 0.1 and 1 μM) or CYT387 (0, 0.1, and 1 μg/ml) overnight. After mechanical shearing, the organoids were incubated with or without 10 μg/ml Poly(I:C), together with the initial concentrations of BX795 or CYT387. At the indicated hours after Poly(I:C) treatment, the organoids were harvested and subjected to RT-qPCR assay to examine mRNA expression levels of bat and human IFNL1 and IFNL3; cell-free media from human organoids were applied to ELISA to detect IFNL1 and IFNL3. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3. Ordinary one-way ANOVA with Tukey’s multiple comparison test. b, c Induction of IFNL1 (left) and IFNL3 (right) in bat intestinal organoids treated with BX795 (b) and CYT387 (c). Results show the fold change of GAPDH-normalized expression level in the Poly(I:C)-treated organoids relative to mock-treated organoids. d, e Induction of IFNL1 and IFNL3 in human intestinal organoids treated with BX795 (d) and CYT387 (e). IFNL1 (f) and IFNL3 (g) secretion from the human organoids at 4 h post-stimulation. h A schematic graph describes the experimental procedure for panels i and j. Bat and human intestinal organoids were pretreated with 1 μg/ml CYT387 or DMSO overnight. After mechanical shearing, the organoids were incubated with 1 μg/ml CYT387 or DMSO with or without 10 μg/ml Poly(I:C) for 4 h. Cell-free media were then harvested and subjected to PRM-MS to analyze bat IFNL3 (i) and ISG15 of bat and human (j). The dotted lines represent the detection limit. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3. Two-tailed unpaired Student’s t test
We developed a parallel reaction monitoring mass spectrometry (PRM-MS) assay for targeted quantification of bat IFNL3 protein and ISG15 protein in the culture medium of bat and human organoids in the presence or absence of CYT387 and Poly(I:C) treatment (Fig. 3h), due to the lack of specific antibodies for detecting bat proteins. The result verified Poly(I:C)-triggered secretion of IFNL3 in bat organoids, which was nullified by CYT387 treatment (Fig. 3i). However, we failed to detect bat IFNL1 by PRM-MS, although the qPCR assay showed a higher induction than IFNL3. The failed identification, we inferred, might be related to the technical issue, rather than the absence of IFNL1 protein in bat organoids. We also measured ISG15 secretion from the bat and human organoids by PRM-MS. Bat ISG15 secreted from Poly(I:C)-treated bat organoids was significantly higher than that in mock-treated organoids, and significantly lower than that in Poly(I:C) + CYT387 double-treated organoids (Fig. 3j, left panel). ISG15 secretion from human organoids was only detectable in Poly(I:C) treated organoids, that in mock-treated organoids and Poly(I:C) + CYT387 double-treated organoids was below the detection limit (Fig. 3j, right panel). Thus, CYT387 treatment substantially dampened bat and human ISG15 production stimulated by Poly(I:C) (Fig. 3j). Here, we had to select specific and distinct peptides to quantify human and bat ISG15, which may result in different detection limits and dynamic ranges for the targeted peptides. Thus, it would be biased to directly compare the abundance of bat and human ISG15, and the magnitude of their induction. Overall, BX795 and CYT387 nullified Poly(I:C)-triggered induction of IFNLs and ISG in both bat and human intestinal organoids, suggesting that TLR3 and RLR signaling might be the conserved pathways mediating antiviral response in bat and human intestinal epithelial cells.
Bat intestinal organoids recapitulating bat susceptibility to viruses
Given that bat intestinal organoids simulated bat intestinal epithelium, we inferred that bat intestinal organoids might be able to reproduce the authentic susceptibility of bat intestinal cells to coronaviruses. Despite the discovery of many bat coronaviruses, only a couple of them have been successfully isolated and cultivated or propagated from molecularly engineered viruses.5,33 A lineage C beta-coronavirus Tylonycteris pachypus coronavirus HKU4 (CoV-HKU4) is one of the cultivable bat coronaviruses, probably owing to its usage of human DPP4 as its cellular receptor.34 In contrast, enterovirus 71 (EV-71) has a limited host range; humans are the only known natural host. We inferred that bat intestinal organoids might be susceptible to CoV-HKU4, but non-susceptible to EV-71. To this end, we tested CoV-HKU4 and EV-71 in bat intestinal organoids. As shown in Fig. 4a, CoV-HKU4 displayed a significantly increased viral gene copy and viral titer after inoculation. Within 48 h post-inoculation, infectious virions in the culture media increased by more than 3 log units. In contrast, we did not observe any sign of viral propagation, no matter whether the bat organoids were exposed to a high or low dose of EV-71 inoculum (Fig. 4b). After a high MOI inoculation (MOI of 1), viral load even decreased gradually after infection, suggesting that bat organoids were non-permissive to the enteric virus of human origin. EV-71 was inoculated in human intestinal organoids as a positive control and displayed a robust viral propagation (Fig. 4c, d), congruent with previous reports.21,26 Notably, CoV-HKU4 also replicated in human intestinal organoids (Fig. 4e), echoing the previous finding of its productive infection in human DPP4 transgenic mice.35 CoV-HKU4 infection in bat and human intestinal organoids was verified by immunofluorescence staining (Fig. 4f) using an antiserum against CoV-HKU4 nucleocapsid protein (NP).
Fig. 4
Bat intestinal organoids reproduced bat susceptibility to coronaviruses. a–c At the indicated hours after bat intestinal organoids inoculated with CoV-HKU4 (a), EV-71 (b) and human intestinal organoids inoculated with EV-71 (c), culture media were harvested and applied to viral load detection by RT-qPCR and viral titration by TCID50 assay. Data represent mean and s.d. in a representative experiment, n = 3. Two-tailed unpaired Student’s t test. d EV-71 infected and mock-infected human intestinal organoids were fixed and immunostained to identify viral protein VP1 positive (green) cells. Nuclei and actin filaments were counterstained with DAPI (blue) and Phalloidin-647 (purple), respectively. Scale bar, 20 µm. e At the indicated hours after human intestinal organoids inoculated with CoV-HKU4, culture media were harvested and applied to viral load detection and viral titration. Data represent mean and s.d. in a representative experiment, n = 3. Two-tailed unpaired Student’s t test. f Bat and human intestinal organoids infected with CoV-HKU4 or mock-infected were fixed and immunostained to identify CoV-HKU4 NP positive (green) cells. Scale bar, 20 µm. g, h At the indicated hours after human and bat intestinal organoids inoculated with WT SARS-CoV-2 and the Omicron variant, culture media were harvested and applied to viral load detection and viral titration. Data represent mean and s.d. in a representative experiment, n = 3. Ordinary one-way ANOVA with Tukey’s multiple comparison test. i SARS-CoV-2 infected and mock-infected bat and human intestinal organoids were fixed and immunolabeled to identify SARS-CoV-2 NP positive (green) cells. Scale bar, 20 µm
RaTG13, a bat coronavirus with 96% homology to SARS-CoV-2, was identified in the droppings of horseshoe bats Rhinolophus affinis.1 Similar to most bat viruses, RaTG13 was not isolated and cultivated. Without live RaTG13 virus, we tested SARS-CoV-2 and found that intestinal organoids derived from horseshoe bats were permissive to SARS-CoV-2 viruses, both the ancestral strain (WT) and the more infectious Omicron variant that emerged in late 2021. Nevertheless, SARS-CoV-2 viruses replicated more actively in human intestinal organoids (Fig. 4g) than in bat organoids (Fig. 4h). Figure 4h showed the replication kinetics in bat organoids after an MOI of 1 inoculation. After inoculation with an MOI of 0.1, the same MOI for inoculating human organoids, the viruses barely replicated in bat organoids (data not shown). Interestingly, the more infectious Omicron variant appeared to possess a higher replicative fitness than the ancestral WT virus in both bat and human organoids. More productive infections of the WT strain and the Omicron variant in human than bat intestinal organoids were verified by immunofluorescence staining (Fig. 4i). Overall, the distinct permissiveness of bat intestinal organoids to a bat coronavirus and an enteric virus of exclusive human origin indicated that bat intestinal organoids recapitulated the authentic susceptibility of live bats to particular viruses. SARS-CoV-2 replication in bat intestinal organoids further supported that bat organoids served as an in vitro correlate of bat intestinal epithelium for phenocopying viral tropism in bat intestines.
We also measured the virus-induced innate immune response in human and bat intestinal organoids. At 48 h post inoculation of SARS-CoV-2, we harvested the human and bat organoids to detect the transactivation of antiviral genes and proinflammatory cytokines. Type III IFNs were highly induced in SARS-CoV-2-infected human organoids (Supplementary Fig. 5), which was consistent to our previous observations24,26 as well as their activation after Poly(I:C) treatment (Fig. 2b). ISGs, TNF-α and IP10 showed a similar induction profile as that in Poly(I:C)-treated human organoids (Supplementary Fig. 5). However, we did not observe a notable induction of these innate immune genes in bat organoids after SARS-CoV-2 infection (data not shown). It, we believe, might be related to the less productive viral replication in bat organoids (Fig. 4g, h).
CYT387 treatment boosting early viral propagation in bat organoids
Now that bat intestinal organoids possessed a higher basal level of antiviral defense than their human counterparts and mounted a more active immune induction upon Poly(I:C) treatment, perturbation of host antiviral defense in bat and human organoids might affect viral growth disparately. Based on the more potent immunosuppressive effect of CYT387 than BX795 as revealed in Fig. 3, we treated bat and human organoids with CYT387, and assessed the impact of blunted immune activation during virus infections. After pretreatment with CYT387 or DMSO overnight, we inoculated bat and human organoids with SARS-CoV-2 and then incubated the organoids with the initial concentrations of CYT387 and DMSO accordingly. We monitored viral growth by detecting the viral load and viral titer in the culture media. As shown in Fig. 5a, CYT387 substantially enhanced viral growth at 8 h post-infection in bat organoids, whereas CYT387 enhancement of viral growth in human organoids occurred at 48 h (Fig. 5b). We performed similar experiments with CoV-HKU4 since both human and bat intestinal organoids sustained CoV-HKU4 replication. The result reproduced a significant boost of viral growth empowered by CYT387 in bat organoids at the early phase of infection (Fig. 5c). However, CYT387-mediated viral enhancement of CoV-HKU4 in human organoids was not as remarkable as that in bat organoids (Fig. 5d). Overall, the results indicated that bat organoids possessed a more robust antiviral machinery enabling instant control of virus replication in the early phase of infection.
Fig. 5
CYT387 treatment enhanced viral replication in bat intestinal organoids in the early stage of viral growth. Bat and human intestinal organoids were pretreated with 1 μg/ml CYT387 or DMSO overnight. After inoculation with WT SARS-CoV-2 or CoV-HKU4, the pretreated bat and human organoids were incubated with 1 μg/ml CYT387 or DMSO. At indicated time points after infection, cell-free media were collected and subjected to viral load detection and viral titration. Data represent mean and s.d. in each organoid line, n = 3. Ordinary one-way ANOVA with Tukey’s multiple comparison test. Viral load and viral titer of SARS-CoV-2 in treated and mock-treated bat organoids (a) and human organoids (b). Viral load and viral titer of CoV-HKU4 in treated and mock-treated bat organoids (c) and human organoids (d)
Source: news.google.com