To compare soil viral community composition within and between terrestrial habitats on a regional scale, viromes were generated from 34 near-surface (top 15 cm) soil samples, with a total of 30 viromes included in ecological analyses. downstream (see Supplementary Methods). The analyzed viromes were collected from four distinct habitats (wetlands, grasslands, chaparral scrub, and forests, each containing 7, 14, 4, and 5 viromes, respectively) at five field sites (Fig. S1 for sampling scheme, Table S1 for soil properties). After quality filtering, the 30 viromes generated an average of 72,950,833 reads and 416 contigs ≥10 Kbp per virome (Table S2). Viromes from wetlands produced more contigs ≥10 Kbp than viromes from other habitats, both in total and on average per virome (Table S2). We used VIBRANT to identify 3,490 viral contigs in our assemblies, which were grouped into 3,432 viral operational taxonomic units (vOTUs), defined as ≥10 Kbp of viral contigs sharing ≥95% average nucleotide identity over 85% contig length. [17]. Restricted principal coordinate analysis (CAP analysis) revealed strong clustering by habitat rather than by site, implying that, where environmental parameters are substantially different, environmental conditions are stronger drivers of viral community composition. than geographical distance (Fig. S2).
Multiple lines of evidence suggest that wetter soils harbored greater viral diversity than drier soils. We recovered the most vOTUs from wetlands, both in total (56% of all vOTUs were from wetlands) and per virome (on average, 307 vOTUs were recovered per wetland virome, compared to 116 from all habitats) (FIG. 1A). As expected, wetlands had significantly higher moisture content than other habitats (Fig. 1B; ANOVA followed by Tukey’s multiple comparisons of means, p< 0.001), especially considering that the soil samples were collected towards the end of the dry season of the Mediterranean climate, which means that most of the habitats had not received precipitation during the previous ~6 months. Although viral richness was higher in wetlands, this was not statistically significant (ANOVA model richness ~ habitat, p= 0.095). Still, nonparametric tests, accounting for nonlinear correlations, revealed a significantly positive correlation between viral richness and soil moisture content (Spearman rho = 0.33, p= 0.036; Kendall’s tau = 0.22, p= 0.045). The beta diversity of the viral community was also related to the chemical properties of the soil in general (Mantel test, R2 = 0.43, p= 0.009; Table S1), while the distance between sites only accounted for 5% of the variation (partial Mantel test, R2 = 0.38, p= 0.009). Overall, viral diversity was generally higher in moist soils.
Fig. 1: Evidence of higher viral richness in wetter soils.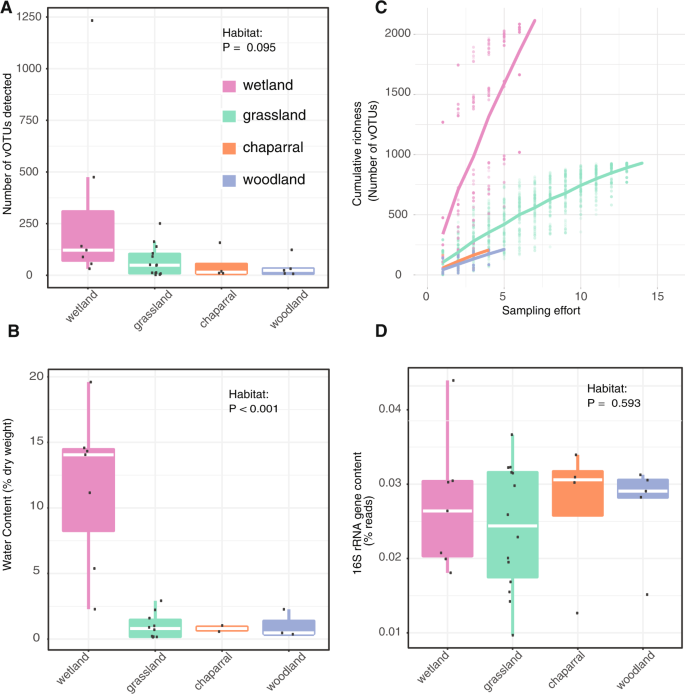
Comparisons between habitats of (A) viral richness (number of identified vOTUs with coverage along at least 75% of the contig in a given virome), the dots represent the richness in individual viromes, (B.) accumulated vOTU richness accumulation curves as sampling effort increases, the points represent the accumulated richness in each sampling effort in 100 permutations of virome order; the overlapping lines show the mean accumulated richness per habitat, (C) the water content, calculated as (wet weight-dry weight) divided by the dry weight, and (D) bacterial 16S rRNA gene content in viromes, based on the percentage of viromic reads mapped to reference 16S rRNA genes. VIBRANT [21] was used to identify 3,490 viral contigs in our assemblies, and these viral contigs clustered with 95% average nucleotide identity (ANI) into 3,432 viral operating taxonomic units (vOTUs). For (A), (B.), (D), the raw data is plotted at the top of the boxplots, with white lines showing the median, boxes indicating 75% of the data, whiskers extending to 90%, and the lowest points beyond the whiskers that indicate outliers.
We then wondered if differences in sampling effort or bacterial content in the viromes could have produced the diversity patterns observed. For example, viral diversity might have appeared artificially higher in wetland habitats if wetland viral diversity was lower but well sampled, compared to other habitats with higher diversity but poorer sampling. In a comparison of the accumulation curves, such a pattern would include a more horizontal slope in the wetlands and a steeper vertical slope over sampling effort in the other habitats. We tested this by comparing accumulation curves between habitats, which revealed the opposite trend: wetlands exhibited the steepest slope increase, and thus were likely to be the least sampled, in terms of true viral diversity (Fig. 1C). . Since relict DNA and small bacteria can pass through 0.22 µm filters, bacterial sequences are known to be present in viromes. [18]. Although we had no specific reason to expect differences in the ratio of bacterial to viral DNA content between habitats, we wanted to exclude the possibility that viral diversity appeared artificially higher in wetlands due to a lower ratio of bacterial content in viromes. of wetlands compared to other habitats. . To compare bacterial content in viromes between habitats, 16S rRNA gene fragments were recovered from raw reads (Fig. 1D). The percentage of 16S rRNA gene sequences in each virome ranged from 0.01 to 0.044% (consistent with previous reports of 0.028% bacterial 16S rRNA gene content in similarly prepared viromes from agricultural soils). [18]) and did not differ significantly by habitat (ANOVA, P= 0.595). Thus, viral diversity estimates did not appear to be disproportionately biased by sampling effort or the presence of non-viral sequences in the viromes. The higher virus diversity in wetlands compared to other drier habitats could be due to higher bacterial activity and thus more susceptible hosts, leading to higher viral activity and diversity. Furthermore, better soil connectivity in moist soils, leading to greater spread of viruses and free hosts, could increase the chances of virus-host encounters.
Perhaps the most surprising result of this study was the uniqueness of each soil viral community. This feature has previously been highlighted in viruses identified in other settings, such as highly oligotrophic waters. [19] and hydraulic fracturing of wells [8]. Most of the vOTUs (81%) in this regional study were only detected in a single virome (Fig. 2A, Fig. S3A). Of the 666 vOTUs detected in at least two viromes, 93% were detected in viromes from the same habitat and site (Fig. 2B, Fig. S3B). The most similar viral communities were from the same habitat and site (ie, biological replicates) less than 1 km apart (Fig. 2C). Within the same site, viral communities were less similar between habitats than within the same habitat. Furthermore, viromes from the same habitat at different nearby sites (within 6 km) did not share any vOTUs, suggesting substantial differences in viral communities along local distances, both within and between habitats. At greater distances, community similarity generally decreased, even between viromes from the same habitat (Fig. 2C). Still, 21 vOTUs were detected in multiple habitats at multiple sites (Fig. 2B), and some vOTUs were shared between the two sites furthest apart (109 km apart, Fig. 2C), suggesting some degree of regional conservation. of viral populations. Overall, the results suggest substantial differences in soil viral community composition in the same habitat at the meter scale, greater similarity of viral communities in close proximity and under similar environmental conditions, and a small number of shared vOTUs. in regional distances.
Fig. 2: Substantial differences in vOTU detection and viral community composition among soil viromes, both within and between habitats.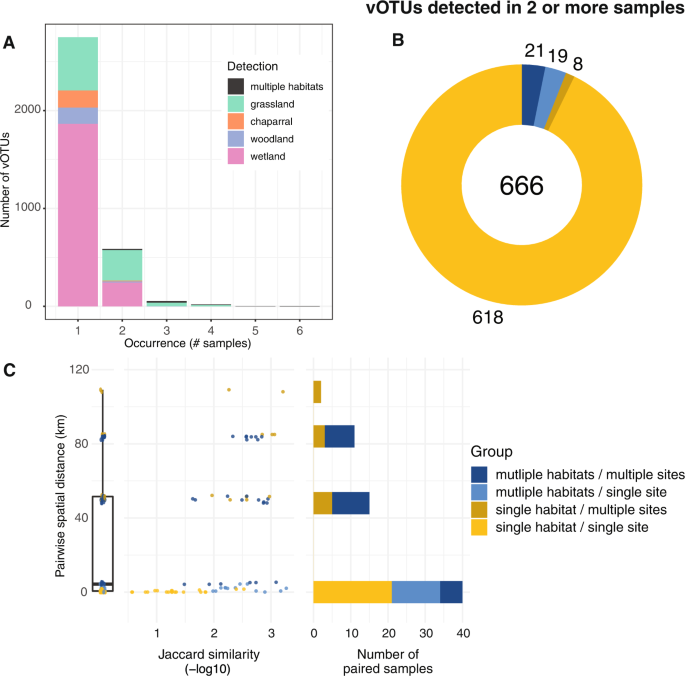
A vOTU detection patterns, in terms of the number of viromes (samples) in which each vOTU was detected (x-axis) and the number of vOTUs showing a given detection pattern (y-axis), with stacked color bars indicating habitats) in which each vOTU was detected, (B.) vOTUs detected in more than one virome, colored by their detection patterns in sites and habitats (legend to the right of panel C), numbers indicate total vOTUs (middle) and vOTUs per detection pattern, and (C) Pairwise community composition similarity (x-axis) by geographic distance (y-axis) between viromes, colored according to detection patterns across habitats and sites (legend at far right). Left plot: boxplot of pairwise distances for all viromes (condensed x-axis). Middle plot: Each point represents the -log10 Jaccard similarity between two viromes along the x-axis, with lower values (left side of the x-axis) indicating greater viral community similarity. Right graph: Total number of virome pairs included in each geographical distance. Paired viromes were only included if they shared at least one vOTU.
Although soil viral communities are woefully undersampled, making sweeping generalizations premature, the results of this study and others converge on a picture of high local viral diversity, with communities differing substantially in space and by habitat, with little regional co-occurrence of viral ‘species’ (UTV). Relationships between distance and viral community decline have been observed in an 18 m long agricultural field [14] and in a meadow of 200 m2 [20], consistent with meter-scale differences in viral community composition between replicates of the same habitat observed in this study. However, the proportion of shared vOTUs in meters varied between these studies, with many vOTUs shared in the agricultural field but most vOTUs not shared between samples separated by ~10 m in this study and in the grassland field, which suggests the potential for different spatial patterns in natural and managed soils, perhaps due to different amounts of soil mixed by management practices, such as tillage. Similarly, substantial differences between viral communities on a regional scale were also identified in a study of RNA viromes from grasslands and peatlands, which shared few viruses between samples. [16]. However, ter Horst et al. showed that 4% of the vOTUs from a peatland in Minnesota, USA were shared across other peatlands, often on different continents [15], consistent with the recovery of a small number of shared vOTUs over distances >100 km here. Collectively, we propose that soil viral communities often display great heterogeneity within and between habitats, presumably due to a combination of host adaptations and microdiversity, dispersal limitation, and fluctuating environmental conditions in space and time.
Source: news.google.com