In a recent study published on the bioRxiv* preprint server, researchers in the United States explored a novel technique to improve representation in viral genomic surveillance.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evaded detection in the early phases of transmission due to a lack of regular viral genomic surveillance, allowing the virus to proliferate unchecked. Furthermore, poor surveillance over the following months allowed SARS-CoV-2 variants to emerge undetected. In light of this, minority groups have experienced a higher burden of coronavirus disease 2019 (COVID-19) cases and deaths due to social and racial inequities.
 Study: a collaborative approach to improve representation in viral genomic surveillance. Image Credit: Studio.c/Shutterstock
Study: a collaborative approach to improve representation in viral genomic surveillance. Image Credit: Studio.c/Shutterstock
About the study
In the present study, the researchers aimed to increase microbial genome sequencing capacity at universities using a new variant surveillance model.
From July 22, 2021 to April 12, 2022, six study partner clinics in Louisiana collected 405 rapid antigen-positive clinical specimens from individuals living in nine boroughs, including Allen, Bienville, Franklin, Jackson, Lincoln, Morehouse , Ouachita, Union, and Webster. More than 90% of the samples contained data related to the demographics and vaccination status of the individuals.
In Georgia, Mercer University School of Medicine (MUSM) samples were collected from Mercer Medicine (MM) community clinics and student health centers in Bibb, Chatham, DeKalb, and Fulton counties. If polymerase chain reaction (PCR) analysis revealed a positive result, samples were sent for Illumina sequencing. The Jackson State University (JSU) Health Services Center in Mississippi served as the collection location. The Center supported the testing and immunization activities of the neighboring community and acted as the primary COVID-19 testing facility for university students, staff, and visitors.
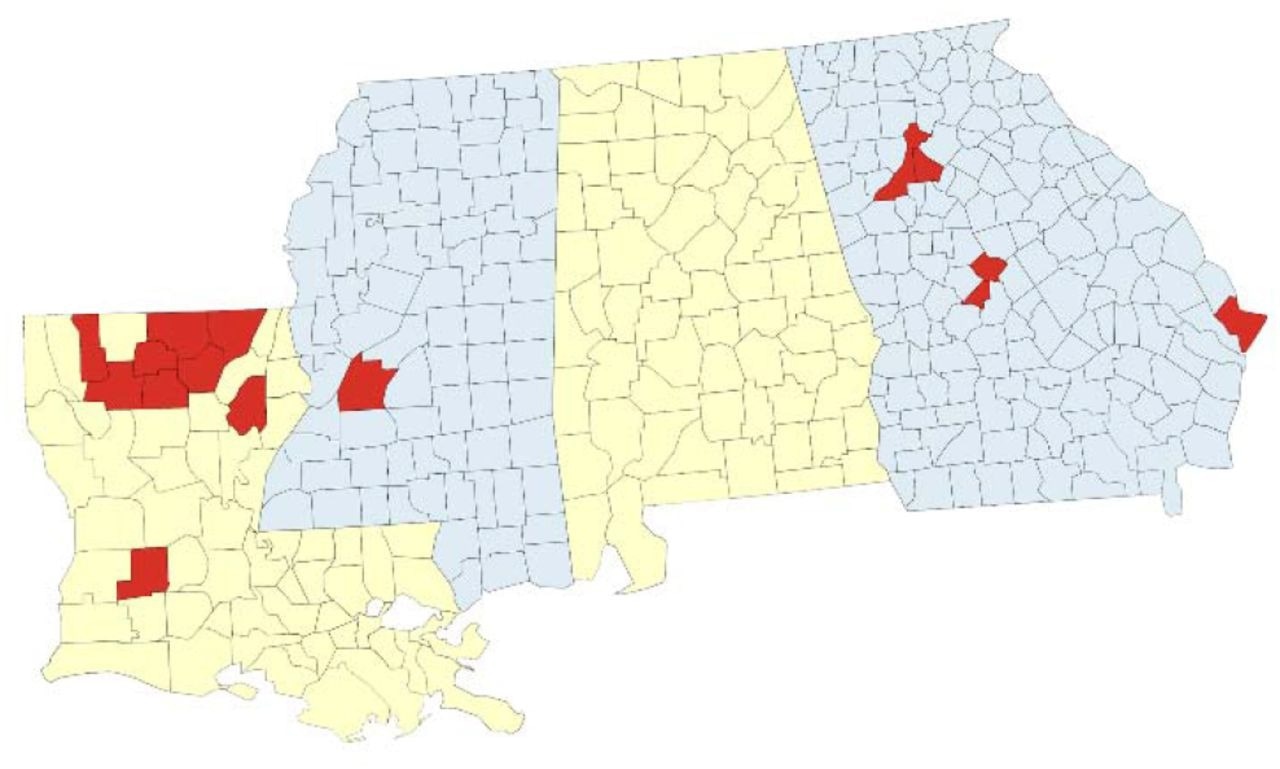 Geographic coverage of SARS-CoV-2 genomic surveillance in Louisiana, Georgia, and Mississippi. Map of the surveillance region with parishes (Louisiana) or counties (Georgia, Mississippi) where at least one specimen was sequenced by the network indicated in red.
Geographic coverage of SARS-CoV-2 genomic surveillance in Louisiana, Georgia, and Mississippi. Map of the surveillance region with parishes (Louisiana) or counties (Georgia, Mississippi) where at least one specimen was sequenced by the network indicated in red.
The team then completed the surveillance cycle by disseminating variant data, weekly wastewater surveillance results, and other COVID-19-related health data through a dashboard created for the people of northern Louisiana. . A matched subset of 16 SARS-CoV-2 positive samples were sequenced to verify the efficiency of Nanopore MinION sequencing.
Results
People who are not white or who are of two or more races accounted for 60% of donors in the study sample compared to 41% for parishes in the sample. Despite representing only 3.1% of the population in northeast Louisiana, Hispanic donors accounted for 8.4% of the samples and 13.8% of the total cases reported by the Louisiana Department of Health. Hispanic people were also significantly more likely to develop COVID-19. Genomes were ultimately recovered from 272 PCR-positive samples out of 405 samples collected in Louisiana, 389 of which showed detectable SARS-CoV-2 ribonucleic acid (RNA).
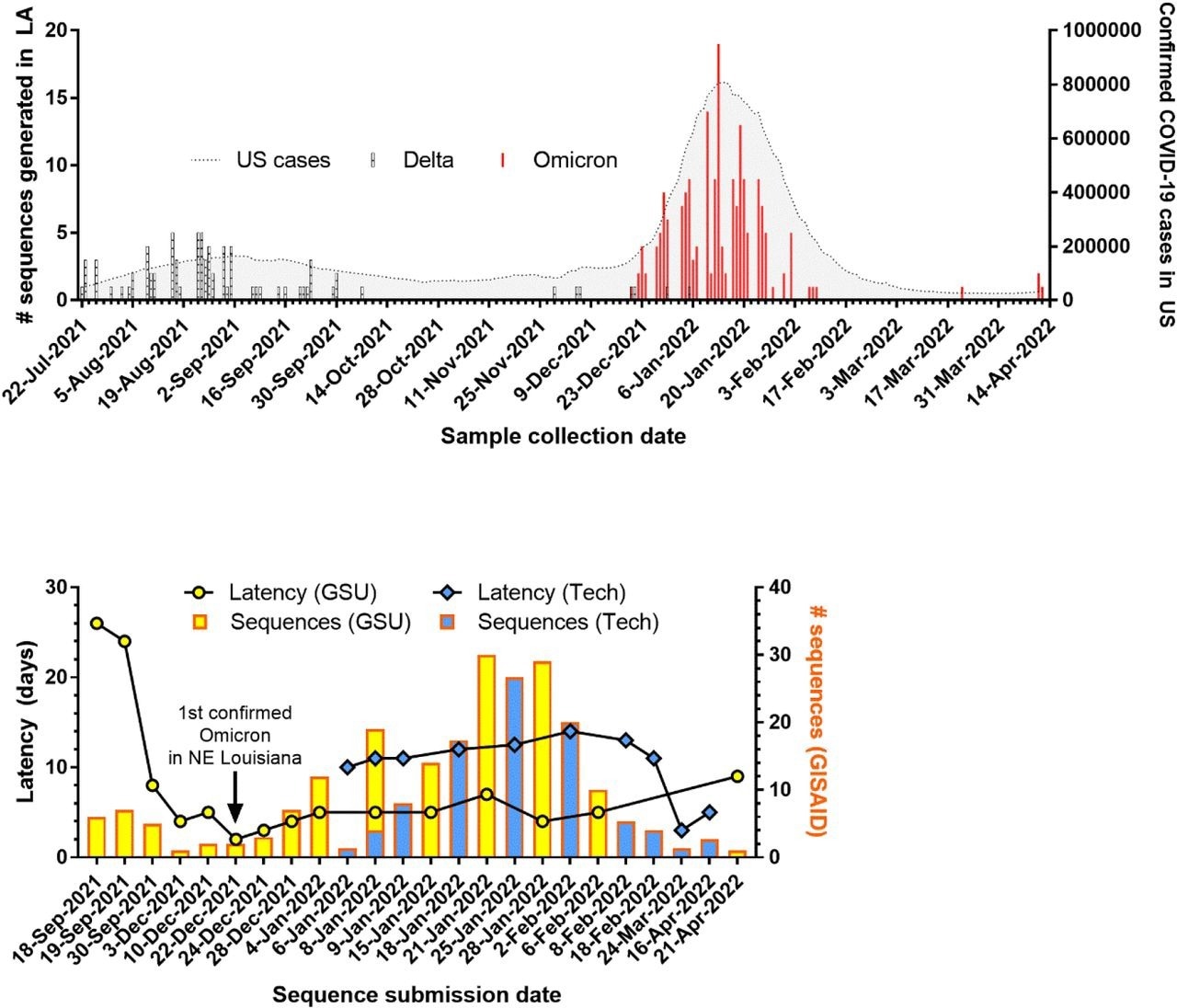 Genomic surveillance of SARS-CoV-2 in Louisiana. (A) Number of sequenced variants over time in Louisiana overlapping with the number of confirmed COVID-19 cases in the US (B) Number of uploaded sequences and latency to new sequencing entities established at Grambling State University (GSU) and Louisiana Tech University (Tech ).
Genomic surveillance of SARS-CoV-2 in Louisiana. (A) Number of sequenced variants over time in Louisiana overlapping with the number of confirmed COVID-19 cases in the US (B) Number of uploaded sequences and latency to new sequencing entities established at Grambling State University (GSU) and Louisiana Tech University (Tech ).
The team discovered two new BA.5 subvariants with immune escape mutations. With 100% consensus sequence identity observed in 13 of the 16 samples along with 99.9% sequence identity in three of the 16 samples, the results demonstrated an efficient precision of the sequencing procedure and analysis process. Nanopore. Within the gene encoding the NSP13 replicase helper protein, all three samples shared the same so-called mismatch base, namely T by Illumina sequencing and G by Nanopore sequencing at position 17,259. The SARS-CoV-2 reference genome also had a G at this location. The evidence of the E1264D mutation found in the samples analyzed with the Illumina methodology was significant. In homopolymeric tracts, the precision of the so-called nanopore base was decreased, but it was not clear why this mismatch was present.
The team found that the mean Cq values of the Omicron specimens were considerably more significant than those of the Delta specimens. This supported previous observations that Omicron infections, unlike Delta infections, had lower viral gene copy numbers, as indicated by real-time reverse transcription PCR (RT-PCR) or infectious viral loads. lowest as determined by the focus formation test. However, this variation was not statistically significant. In addition, the mean Cq value of unvaccinated individuals with no prior disease was 1.06 cycles lower than that of fully vaccinated individuals with or without a history of prior infection.
Overall, the study findings showed that fostering collaborations between academic teams and neighborhood health clinics and instituting genomic surveillance capacity in rural academic institutions resulted in an effective strategy for improving pandemic preparedness.
*Important news
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and therefore should not be considered conclusive, guide clinical practice or health-related behavior, or treated as established information.
Magazine reference:
A Collaborative Approach to Improving Representation in Viral Genomic Surveillance, Paul Kim, Audrey Y. Kim, Jamie J. Newman, Eleonora Cella, Thomas C. Bishop, Peter J. Huwe, Olga N. Uchakina, Robert J. McKallip, Vance L. Mack, Marnie P. Hill, Ifedayo Victor Ogungbe, Olawale Adeyinka, Samuel Jones, Gregory Ware, Jennifer L. Carroll, Jarrod F. Sawyer, Kenneth H. Densmore, Michael Foster, Lescia Valmond, John Thomas, Taj Azarian, Krista Queen, Jeremy P. Kamil, bioRxiv 2022.10.19.512816, DOI: https://doi.org/10.1101/2022.10.19.512816, https://www.biorxiv.org/content/10.1101/2022.10.19.512816v2
Source: news.google.com