Comparison of viral RNA shedding following inoculation of ferrets with IAVs displaying diverse transmissibility profiles
The 2009 A(H1N1)pdm09 influenza pandemic highlighted the potential of swine-origin influenza viruses to cause human disease and global spread. Since then, there has been an increased focus on IAV surveillance and mammalian transmissibility assessments using the ferret model. Swine-origin influenza viruses typically transmit easily between cohoused ferrets but differ widely in their ability to transmit between ferrets in setting where direct and indirect contact (fomites on intermediate surfaces) are eliminated, and transmission can only occur via air7,9. Unfortunately, routine ferret transmission studies do not include assessments of the kinetics and levels of virus released into the air by infected animals. To address this gap in knowledge we analyzed virus levels in nasal washes concurrent with quantification of viral RNA levels in the air. We selected three H1 subtype swine-origin influenza viruses which were previously shown to transmit between ferrets in a direct contact setting but displayed differences in transmission via air: A/Minnesota/45/2016 (MN/45) A(H1N2)v virus transmitted between 3/3 ferret pairs, A/Ohio/09/2015 (OH/09) A(H1N1)v virus transmitted between 1/3 ferret pairs, while A/Ohio/02/2007 (OH/02) A(H1N1)v virus did not transmit via air between any of the ferret pairs (Table 1)25,26,27. To evaluate airborne virus shedding three ferrets per group were inoculated with each of the 3 viruses. Nasal washes were collected from inoculated ferrets every other day for 9 days post-inoculation (p.i.) and were analyzed to determine the levels of infectious virus and viral RNA. Simultaneously, aerosols were sampled from ferret cages for 2 h/day on days 1, 2, 3, and 5 p.i. using a two-stage cyclone aerosol sampler (BC 251 model) that separates particles based on size into three fractions: >4 µm, 4–1 µm, and <1 µm24. Infectious virus was detected in nasal wash specimens for all viruses up to day 5 p.i., while viral RNA was detected through day 9 p.i. Viral RNA copy number averages were 1.9–3.7 orders of magnitude higher than average infectious virus titers detected between days 1 and 5 p.i. Comparable mean peak nasal wash titers were observed for MN/45 and OH/09 viruses (7.0 and 6.9 log10 PFU/mL, and 9.2 and 8.9 log10 RNA copies/mL, respectively), both of which were higher than observed peak titers for OH/02 virus (5.3 log10 PFU/ml and 8.1 log10 RNA copies/mL) (Fig. 1a, c, e). Despite high viral loads in the upper respiratory tract as measured in ferret nasal wash samples on day 1 p.i., negligible levels of viral RNA were detected in the air on day 1 p.i. (Fig. 1b, d. f). However, by day 2 p.i., high levels of viral RNA were detected in the air from all ferrets inoculated with MN/45 and OH/09 viruses. The viral RNA was collected primarily in the >4 µm fraction and was detectable for most ferrets through day 5 p.i. In contrast, the OH/02 virus RNA was not elevated in air samples until day 5 p.i. To determine whether the differences in replication kinetics and release of viral RNA into the air were statistically significant between the viruses tested, we compared area under the curve values for viral RNA copies detected in the nasal wash specimens and in the air collected through day 5 p.i. The results revealed that the viral RNA copy numbers in nasal washes as well as in the air samples collected from ferrets infected by OH/02 virus were significantly lower than for those infected by MN/45 virus, a highly transmissible virus (p = 0.02) (Fig. 1g, h). Collectively, these data show that collection of virus-laden aerosols shed by influenza virus-infected ferrets can provide meaningful contextual information regarding transmission capability, especially when collected early after inoculation.
Table 1 Transmissibility profiles of IAVs evaluated in this study.Fig. 1: Influenza A viral RNA measured in the air following inoculation of ferrets.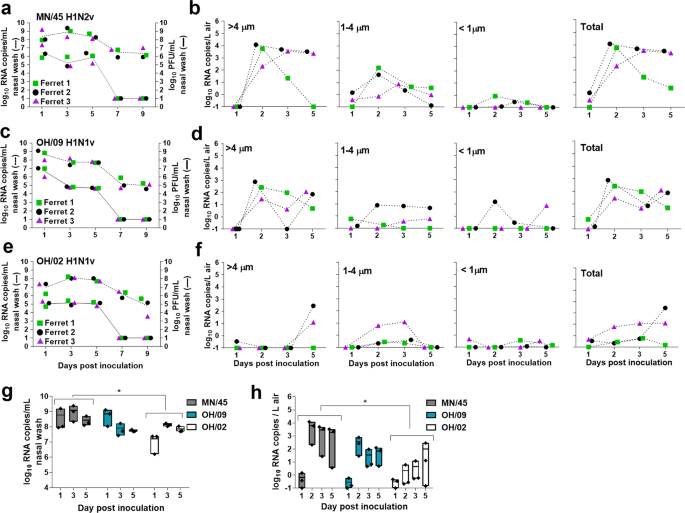
Nasal wash specimens from ferrets (n = 3 per virus) inoculated with A/Minnesota/45/2016 A(H1N2)v a, A/Ohio/09/2015 A(H1N1)v c, and A/Ohio/02/2007 A(H1N1)v e were collected on days 1, 3, 5, 7, 9 p.i. Infectious virus was quantified by standard plaque assay (solid lines) and viral RNA copies were quantified using real time qRT-PCR (dashed lines). Individual ferrets are shown as different symbols (ferret 1-green square, ferret 2-black circle, ferret 3-purple triangle). Limit of detection was 1 log10 PFU/ml or RNA copies/ml of nasal wash. Air samples were collected from each ferret cage for 2 hours on indicated days using a two-stage cyclone sampler which fractioned particles by size: >4 µm, 1–4 µm, <1 µm. Viral RNA in each sample was quantified using real time qRT-PCR and the data for each virus; A/Minnesota/45/2016 A(H1N2)v b, A/Ohio/09/2015 A(H1N1)v d, A/Ohio/02/2007 A(H1N1)v f, and individual ferret is shown. Limit of detection was 21 copies per 210 liters of air collected at each time point (0.1 RNA copy/L of air). Summary figures comparing viral RNA copies detected in the nasal wash specimens g and in the air h collected through day 5 p.i. The points on graphs are the data for individual ferrets, the bars represent minimum and maximum, and the lines represents the mean. Area under the curve for each of these time courses was calculated and statistical significance was evaluated using One-Way ANOVA (Tukey’s post-test [g]) and Kruskal-Wallis test (Dunn’s post-test [h]). *p < 0.05.
Quantification of airborne viral RNA following inoculation of ferrets with IAVs that transmit via air
Next, we sought to quantify the levels of viral RNA released into the air by inoculated ferrets that were housed in cages adjacent to contact ferrets during experiments utilizing the RDT model. Perforated sidewalls adjoining the cages were blocked during air collection to prevent collecting air from the contact ferrets. Viruses examined included three human seasonal influenza viruses [A/Idaho/07/2018 (ID/07) A(H1N1)pdm09, A/Michigan/45/2015 (MI/45) A(H1N1)pdm09, A/Nebraska/14/2019 (NE/14) A(H1N1)pdm09], three swine-origin influenza viruses [A/Michigan/288/2019 (MI/288) A(H1N1)v, A/California/62/2018 (CA/62) A(H1N2)v, A/Ohio/24/2017 (OH/24) A(H1N2)v], and one low pathogenic avian influenza (LPAI) virus [A/Anhui-Lujiang/39/2018 (Anhui-Lujiang/39) A(H9N2)]. Detailed information about clinical signs of infection as well as transmission and pathogenesis data can be found in references provided in Table 1. ID/07, MI/45, NE/14, and MI/288 viruses transmitted efficiently through the air between 3/3 ferret pairs by day 5 post-contact, Anhui-Lujiang/39 virus transmitted by day 3 p.c. between 2/3 ferret pairs, while CA/62 and OH/24 viruses transmitted by day 3 p.c. between 1/3 ferret pairs as evidenced by detection of infectious virus in nasal wash samples5,28. Viral RNA copy levels in nasal washes from animals inoculated with viruses that transmitted efficiently through the air (ID/7, MI/45, NE/14, and MI/288 peaked on day 1 p.i. and ranged between 9.3 and 10.0 log10 RNA copies/mL, which was 2–3 orders of magnitude higher than infectious virus (Fig. 2a, c, e, g). Viruses that transmitted inefficiently via air between ferrets (CA/62, OH/24, and Anhui-Lujiang/39) peaked variably between day 1 and 5 p.i., and the mean peak titers were 8.2–8.5 log10 RNA copies/mL (Fig. 2i, k, m).
Fig. 2: Influenza A viral RNA measured in the air during ferret transmission experiments.
Three ferrets per group were inoculated with A/Idaho/07/2018 A(H1N1)pdm09 a, A/Michigan/45/2015 A(H1N1)pdm09 c, A/Nebraska/14/2019 A(H1N1)pdm09 e, A/Michigan/288/2019 A(H1N1)v g, A/Anhui-Lujiang/39/2018 A(H9N2) i, A/California/62/2018 A(H1N2)v k, A/Ohio/24/2017 A(H1N2)v m. The following day a naïve ferret was placed in an adjacent cage allowing for air exchange via perforated cage walls to assess airborne transmission. Infectious virus in nasal wash samples was quantified using plaque assay or by egg titration (solid lines; data source listed in Table 1; limit of detection 1 log10 PFU/ml or 1.5 log10 EID50/ml), and viral RNA copies were quantified using real time qRT-PCR (dashed lines; data generated in this study; limit of detection 1 log10 RNA copies/ml). Individual ferrets are shown as different symbols (ferret 1-green square, ferret 2-black circle, ferret 3-purple triangle). The plus sign indicates that infectious virus was detected in nasal wash samples collected from contact animals by day 5 post-contact; the negative sign indicates that transmission did not occur as confirmed by lack of the virus in nasal wash samples and by lack of seroconversion (Transmission data source listed in Table 1). Air samples were collected from each ferret cage for 2 h on indicated days using a two-stage cyclone sampler which fractioned particles by size: >4 µm, 1–4 µm, <1 µm (perforated cage side wall was blocked during air sampling). Viral RNA in each sample was quantified using real time qRT-PCR and the data for each virus; A/Idaho/07/2018 A(H1N1)pdm09 b, A/Michigan/45/2015 A(H1N1)pdm09 d, A/Nebraska/14/2019 A(H1N1)pdm09 f, A/Michigan/288/2019 A(H1N1)v h, A/Anhui-Lujiang/39/2018 A(H9N2) j, A/California/62/2018 A(H1N2)v l, A/Ohio/24/2017 A(H1N2)v n, and individual ferrets is shown (limit of detection 0.1 RNA copy/L of air).
For most ferrets inoculated with ID/7, MI/45, NE/14 or MI/288 virus, airborne viral RNA was detected at quantities ≥2 log10 RNA copies/L of air starting on day 1 p.i. and was maintained at 2-4 log10 RNA copies/L on days 2 and 3 p.i. (Fig. 2b, d, f, h). Animals inoculated with the Anhui-Lujiang/39 virus or CA/62 virus released negligible quantities of viral RNA into the air on day 1 p.i. (Fig. 2j, l). The two ferrets (Ferret 2 and 3) inoculated with Anhui-Lujiang/39 virus that released 3–4 log10 RNA copies/L on days 2–5 p.i. transmitted virus to their respective contact ferrets. Ferret 1, which released <1 log10 RNA copies/L on days 1–3 p.i., and 5.3 log10 RNA copies/L on day 5 p.i., did not transmit virus to the contact animal. Transmission was not observed from donor ferrets 1 and 3 that were inoculated with CA/62 virus and released ≤1 log10 RNA copies/L on days 1–5, while transmission did occur for ferret 2 that released 1–2 log10 RNA copies/L on days 2–5. The OH/24-inoculated ferrets released high levels of viral RNA (up to 5.4 log10 RNA copies/L); however, transmission occurred only between one pair (Ferret 2, Fig. 2n), highlighting that other factors, in addition to the amount of airborne virus, play an important role in the transmission of zoonotic influenza viruses. Collectively, these data show that the animals releasing high quantities of viral RNA in the air at early time points post-inoculation were most frequently associated with transmission events to contact ferrets, whereas animals that released low levels of viral RNA in the air or that shed peak levels at later time points post-inoculation, were less likely to transmit virus to contact ferrets in this setting.
Quantification of airborne viral RNA following inoculation of ferrets with IAVs that poorly transmit
To further expand our analysis, we next evaluated a panel of viruses that do not transmit or poorly transmit in the ferret model. As reported elsewhere (Table 1), the highly pathogenic avian influenza (HPAI) A/Yunnan/14563/2015 (Yunnan/14563) A(H5N6) virus did not transmit between co-housed ferrets, while the LPAI A/turkey/California/18-031151-4/2018 (tr/CA/18-031151-4) A(H7N3) and HPAI A/Sichuan/26221/2014 (Sichuan/26221) and A/duck/Bangladesh/19D770/2017 (dk/Bang/19D770) A(H5N6) viruses transmitted between 1/3 co-housed contact animals29,30. To investigate if reduced shedding of airborne viral RNA early after infection coincided with the lack of robust transmission among ferrets, we collected air from inoculated ferrets on days 1-3 p.i. (the period identified as most critical for transmission, Fig. 2). Out of the four viruses tested, ferrets inoculated with tr/CA/18-031151-4 virus displayed the lowest nasal wash RNA titers (5.9 log10 copies/mL) and released negligible levels of viral RNA into the air at ≤1.2 log10 copies/L on day 3 p.i. in the >4 µm fraction and <1 log10 copies/L in the other fractions (Fig. 3a, b). Ferrets inoculated with Sichuan/26221 virus also showed a delay in viral RNA release into the air with total titers up to 2.2 log10 RNA copies/L observed on day 3 p.i., which coincided with increased nasal wash titers observed at this later time point (mean peak of 7.9 log10 RNA copies/mL) (Fig. 3c, d). In the case of Yunnan/14563 virus-inoculated ferrets, 2/3 ferrets released 0.5–2.2 log10 RNA copies/L of air in the >4 µm fraction while the third ferret shed viral RNA in other fractions. Despite the differences in Yunnan/14563 RNA release into the air, comparable nasal wash titers were observed for the three ferrets with a mean peak at 7.4 log10 RNA copies/mL (Fig. 3e, f). Each of the three dk/Bang/19D770 virus-inoculated ferrets released detectable viral RNA into the air (≤2.0 log10 copies/L) by day 1 p.i.; however, only one of the ferrets maintained viral RNA shedding through days 2 and 3 p.i., while mean peak viral RNA levels in nasal wash were 6.4 log10 copies/ml from these ferrets (Fig. 3g, h). Unlike what was observed for the transmissible viruses, the avian strains tested here displayed less robust replication in the upper respiratory tract of ferrets, delayed replication kinetics, with low levels of viral RNA were released into the air within the first three days post-inoculation.
Fig. 3: Quantification of airborne viral RNA released from ferrets inoculated with poorly transmissible influenza A viruses.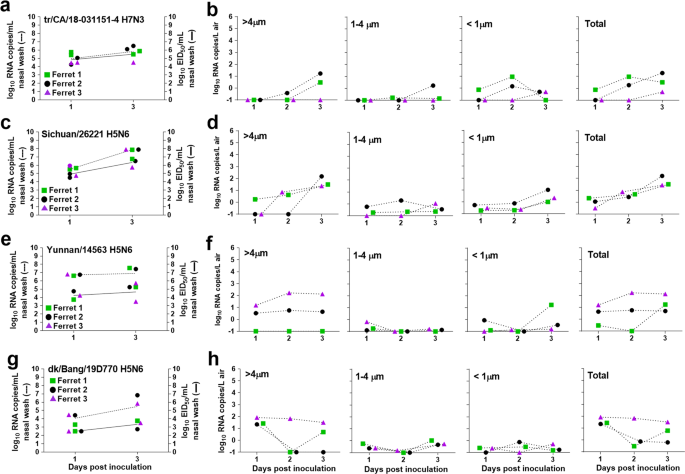
Three ferrets per group were inoculated with A/turkey/California/18-031151-4/2018 A(H7N3) a, A/Sichuan/26221/2014 A(H5N6) c, A/Yunnan/14563/2015 A(H5N6) e, and A/duck/Bangladesh/19D770/2017 A(H5N6) g. Infectious virus in nasal wash samples was quantified by egg titration (solid lines; data source listed in Table 1; limit of detection 1 log10 EID50/ml), and viral RNA copies were quantified using real time qRT-PCR (dashed lines; data generated in this study; limit of detection 1 log10 RNA copies/ml). Each individual ferret is shown as a different symbol (ferret 1-green square, ferret 2-black circle, ferret 3-purple triangle). Air samples were collected from each ferret cage for 2 h on indicated days using a two-stage cyclone sampler which separated the aerosol particles based on size into three fractions: > 4 µm, 1–4 µm, <1 µm. Viral RNA in each sample was quantified using real time qRT-PCR and the data for each virus; A/turkey/California/18-031151-4/2018 A(H7N3) b, A/Sichuan/26221/2014 A(H5N6) d, A/Yunnan/14563/2015 A(H5N6) f, and A/duck/Bangladesh/19D770/2017 A(H5N6) h, and the individual ferret is shown (limit of detection 0.1 RNA copy/L of air).
Comparative analysis
Previous studies have shown that the frequency of transmission through the air to a contact ferret decreases with time underscoring the importance of robust virus replication at early time points after inoculation15,31,32. To determine whether there is a significant difference in nasal wash titers collected at early times points p.i. (day 1, day 3 or peak titer) between transmissible and non-transmissible viruses we classified the viruses in three groups based on their transmissibility profile and performed statistical analyses. The results revealed that regardless of time point analyzed, viral RNA levels in nasal washes collected from ferrets inoculated with viruses that transmitted with 100% frequency though the air (transmission between 3/3 ferret pairs; ID/07, MI/45, NE/14, MI/288, MN/45) were significantly higher when compared to the titers detected from ferrets inoculated with viruses that transmitted less efficiently (Fig. 4). The levels of RNA in nasal wash samples collected on both day 1 and 3 p.i. from ferrets inoculated with viruses that transmitted with 66-33% frequency (transmission between 2/4 or 1/3 ferret pairs; Anhui-Lujiang/39, CA/62, OH/24, OH/09) were also significantly higher than titers detected from ferrets inoculated with viruses in the 0% transmission efficiency group (OH/02, tr/CA/18-031151-4, Sichuan/26221, Yunnan/14563, dk/Bang/19D770).
Fig. 4: Comparison of viral RNA levels detected in nasal wash samples from ferrets inoculated with influenza A viruses displaying diverse transmission capabilities.
Viral RNA copies in nasal wash samples collected from inoculated ferrets on day 1 p.i. a, day 3 p.i. b, or peak titer (either d1 or d3 p.i.) c are shown. Data were grouped by virus transmissibility profile in the ferret model (100%-transmission between all ferret pairs [A/Idaho/07/2018 A(H1N1)pdm09, A/Michigan/45/2015 A(H1N1)pdm09, A/Nebraska/14/2019 A(H1N1)pdm09, A/Michigan/288/2019 A(H1N1)v, A/Minnesota/45/2016 A(H1N2)v (3 ferrets per virus; n = 15)], 66–33% – transmission between 2/3 or 1/3 ferret pairs [A/Anhui-Lujiang/39/2018 A(H9N2), A/California/62/2018 A(H1N2)v, A/Ohio/24/2017 A(H1N2)v, A/Ohio/09/2015 A(H1N1)v (3 ferrets per virus; n = 12)], 0% – no transmission via air or inefficient direct contact transmission if airborne transmission was not tested [A/Ohio/02/2007 A(H1N1)v, A/turkey/California/18-031151-4/2018 A(H7N3), A/Sichuan/26221/2014 A(H5N6), A/Yunnan/14563/2015 A(H5N6), A/duck/Bangladesh/19D770/2017 A(H5N6) (3 ferrets per virus; n = 15)]. Kruskal–Wallis one-way analysis of variance (ANOVA) with Dunn’s post-test were used to determine significant differences, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data points represent individual ferrets. Error bars represent median with interquartile range.
Next, we similarly analyzed the data obtained from air samples. Consistent with nasal wash RNA titer data, ferrets infected with transmissible viruses released significantly more viral RNA into the air than non-transmissible viruses at early timepoints p.i. (Fig. 5). The differences were more pronounced for viral particles collected in the >4 µm fraction, though some of these trends were also observed for particles 1–4 µm in size. In contrast, the viruses that displayed no or limited transmission were found at significantly higher levels in <1 µm fractions collected on day 3 p.i., indicating that this group of viruses may be aerosolized at later time points after inoculation from different parts of the ferret respiratory tract. Nonetheless, the overall levels of viral RNA in these fractions were much smaller as compared to other fractions. These data indicate that transmissible viruses have an upper respiratory tract replication advantage, thus achieving high replication peaks faster and releasing higher quantities of virus into the air, while viruses that are not capable of transmission via the air, in general, display delayed and lower nasal wash titers and peak titers in air samples.
Fig. 5: Detection of influenza A virus genomic RNA in size-fractioned air samples collected from ferrets.
Viral RNA copies in particle size-fractionated air samples collected from inoculated ferrets on days 1-3 and peak viral RNA copies are shown (total RNA is inclusive of all size fractions). Viruses were grouped by transmissibility profile in the ferret model (100%-transmission between all ferret pairs [A/Idaho/07/2018 A(H1N1)pdm09, A/Michigan/45/2015 A(H1N1)pdm09, A/Nebraska/14/2019 A(H1N1)pdm09, A/Michigan/288/2019 A(H1N1)v, A/Minnesota/45/2016 A(H1N2)v (3 ferrets per virus; n = 15)], 66–33%—transmission between 1/3 or 2/3 ferret pairs [A/Anhui-Lujiang/39/2018 A(H9N2), A/California/62/2018 A(H1N2)v, A/Ohio/24/2017 A(H1N2)v, A/Ohio/09/2015 A(H1N1)v (3 ferrets per virus; n = 12)], 0%—no transmission via air or inefficient direct contact transmission if airborne transmission was not tested [A/Ohio/02/2007 A(H1N1)v, A/turkey/California/18-031151-4/2018 A(H7N3), A/Sichuan/26221/2014 A(H5N6), A/Yunnan/14563/2015 A(H5N6), A/duck/Bangladesh/19D770/2017 A(H5N6) (3 ferrets per virus; n = 15)]. Kruskal–Wallis one-way analysis of variance (ANOVA) with Dunn’s post-test was used to determine significant differences, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data points represent individual ferrets. Error bars represent the median with interquartile range.
Source: news.google.com